AlphaFold2源码解析(6)--模型之特征表征
创始人
2024-03-14 16:36:49
0次
AlphaFold2源码解析(6)–模型之特征表征
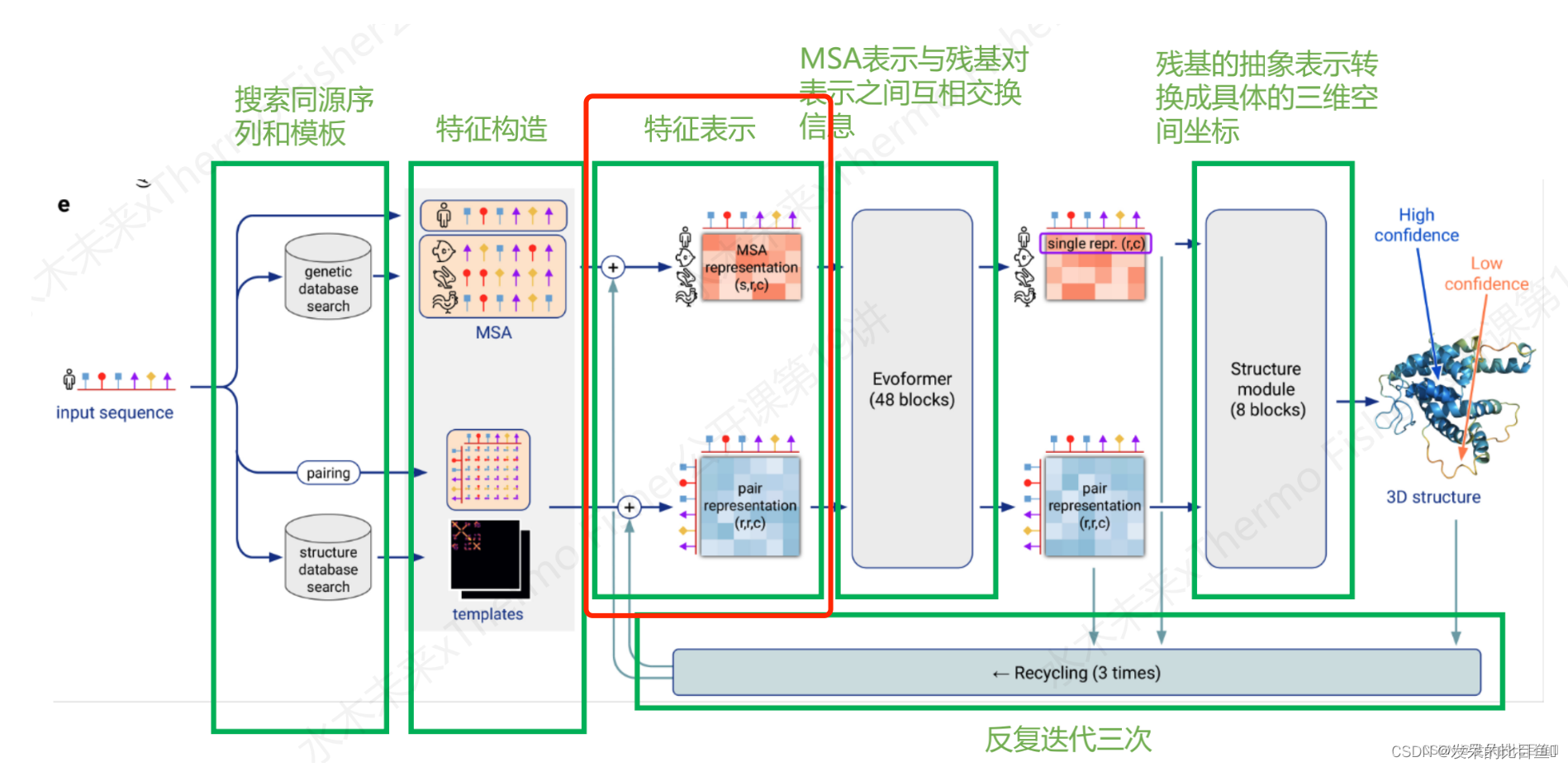 整体推理说明:
整体推理说明:
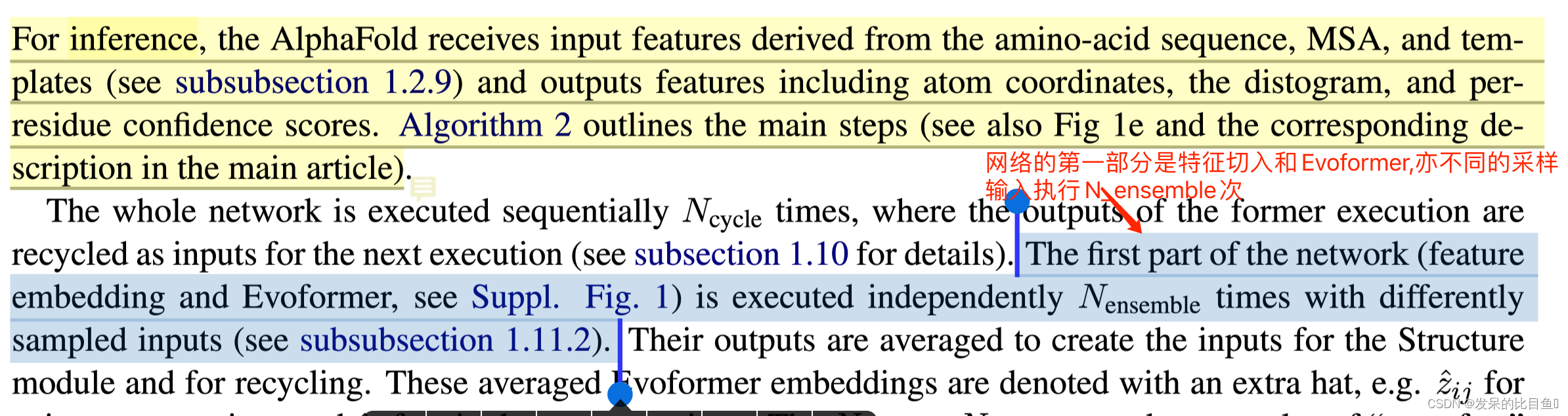
Embedding只是在推理使用,影响非常小(sup-Inference篇章)

特征表征表示的入口模型如下:
evoformer_module = EmbeddingsAndEvoformer(self.config.embeddings_and_evoformer, self.global_config)
其中:
embeddings_and_evoformer是模型的配置参数
self.config.embeddings_and_evoformer.keys()
['evoformer', 'evoformer_num_block', 'extra_msa_channel', 'extra_msa_stack_num_block', 'max_relative_feature', 'msa_channel', 'pair_channel', 'prev_pos', 'recycle_features', 'recycle_pos', 'seq_channel', 'template']
global_config全局配置参数
self.global_config.keys()
['deterministic', 'multimer_mode', 'subbatch_size', 'use_remat', 'zero_init']
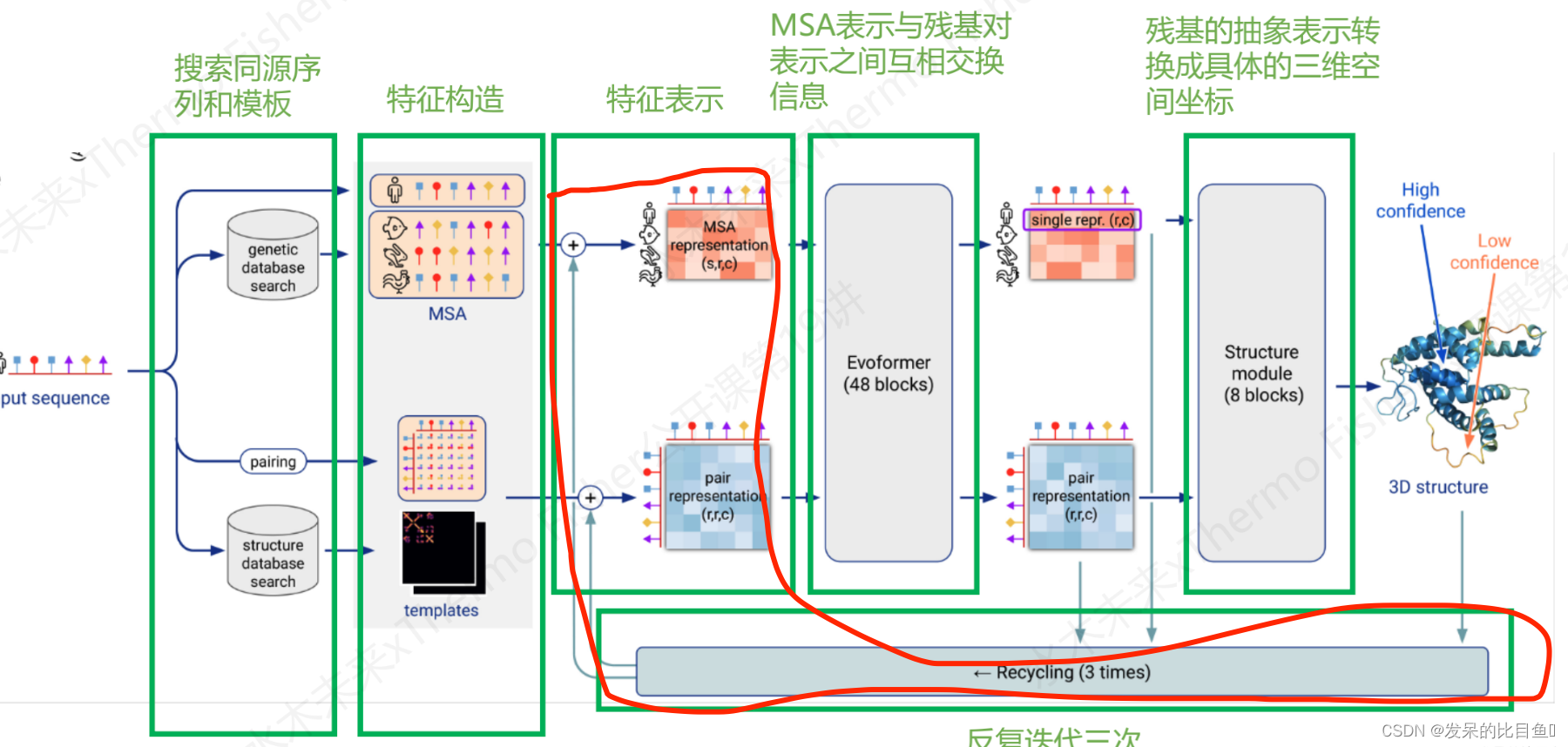
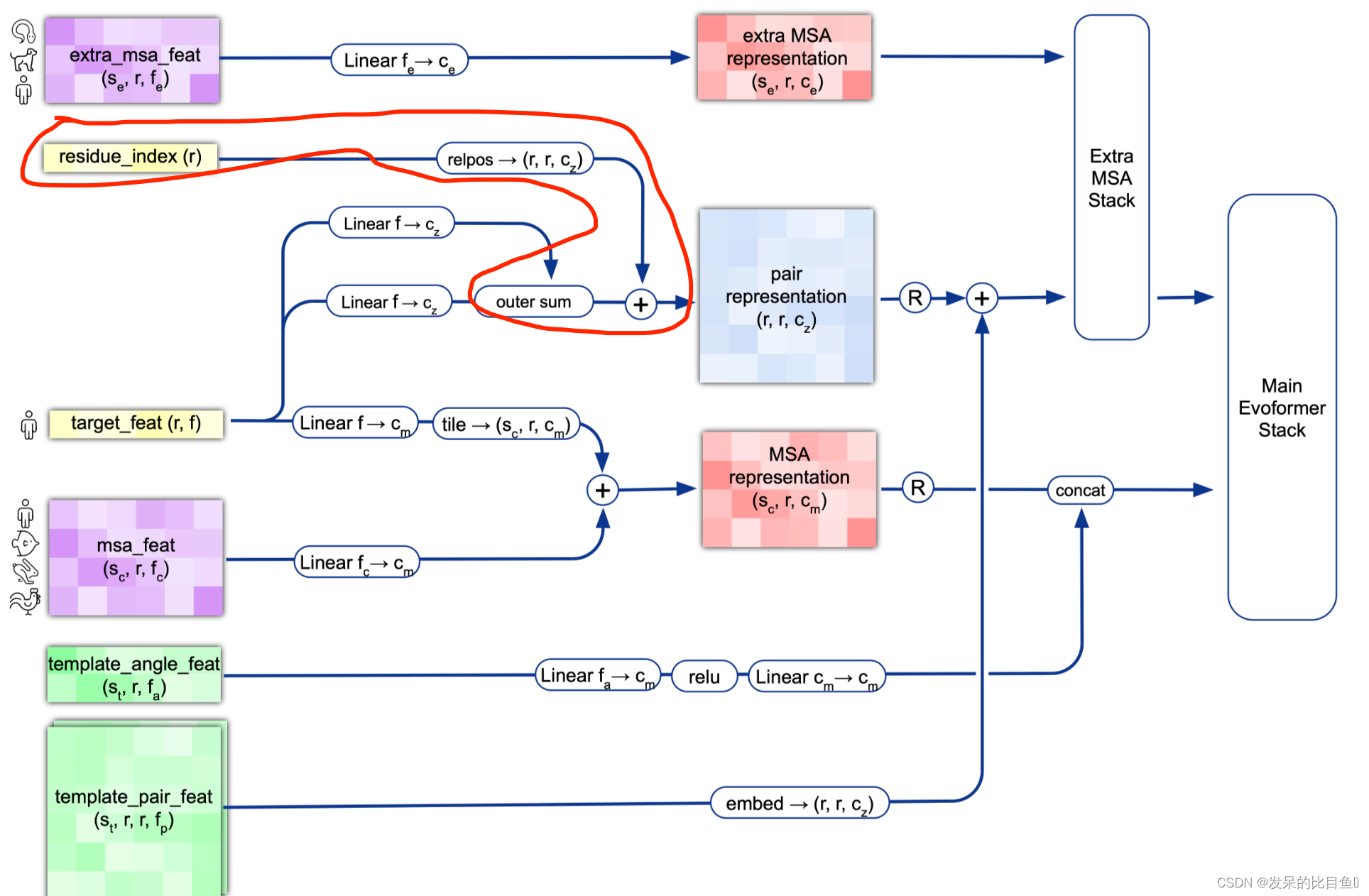
整体Embedding流程
target_feat: shape(N_res, 21) 一个由aatype特征组成residue_index: shape(N_res), 由residue_index特征组成。msa_feat:shape(N_clust, N_res, 49)的特征,由cluster_msa、cluster_has_deletion、cluster_deletion_value、cluster_deletion_mean、cluster_profile拼接而成。extra_msa_feat: shape(N_extra_seq, N_res, 25)的特征,由extra_msa、extra_msa_has_deletion、extra_msa_deletion_value连接而成。与上面的msa_feat一起,还从这个特征中抽取N_cycle×N_ensemble随机样本template_pair_feat: shape(N_templ, N_res, N_res, 88), 由template_distogram和template_unit_vector组成,template_aatype特征是通过平铺和堆叠包含的(这在两个残基方向上完成了两次)。还包括掩码特征template_pseudo_beta_mask和template_backbone_frame_mask,其中特征f_ij=mas_ki·mas_kj。template_angle_feat: shape(N_templ, N_res, 51)特征,由template_aatype,template_torsion_angles,template_alt_torsion_angles, 和template_torsion_mask组成。
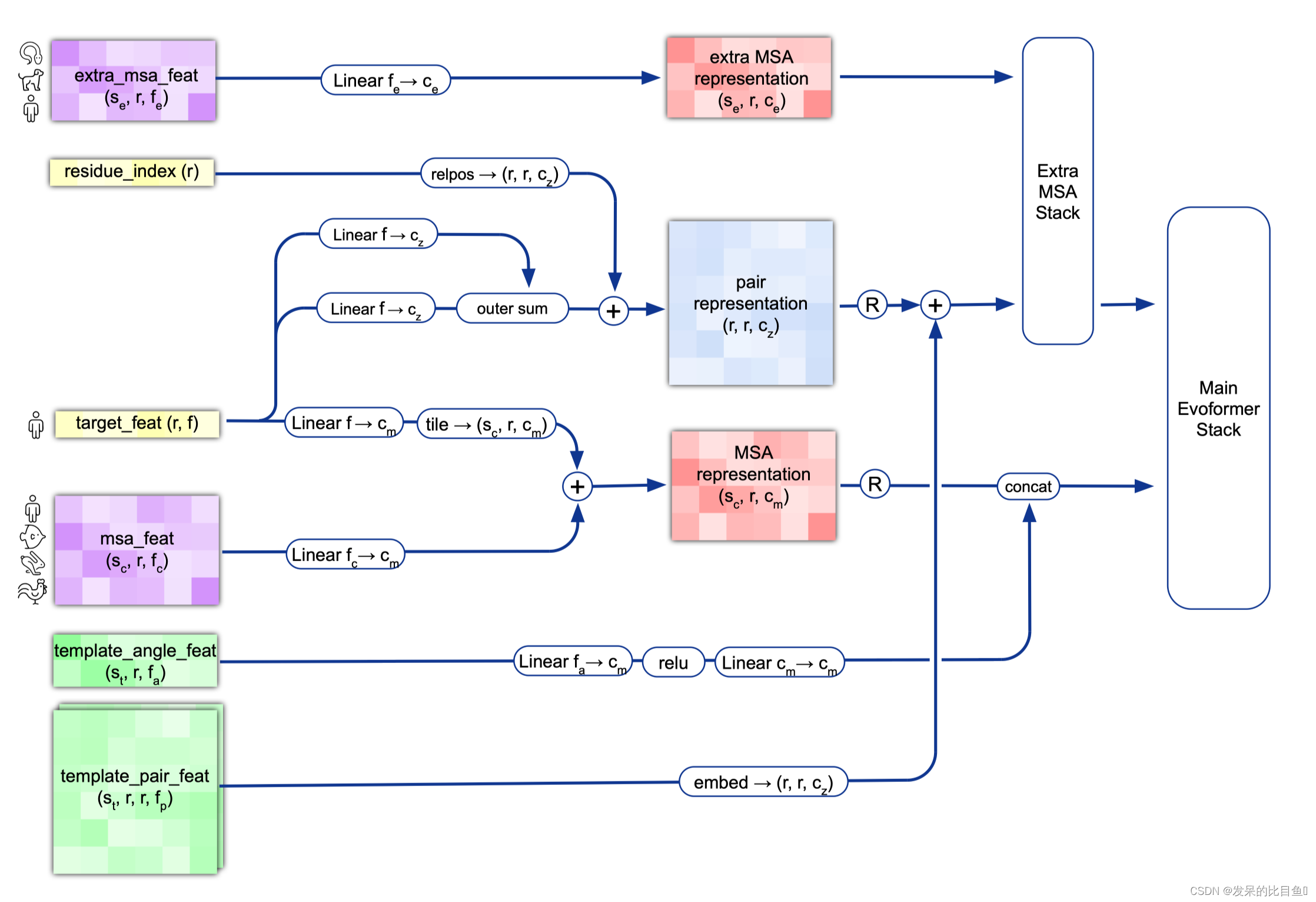
内容翻译如下:
MSA Embedding:
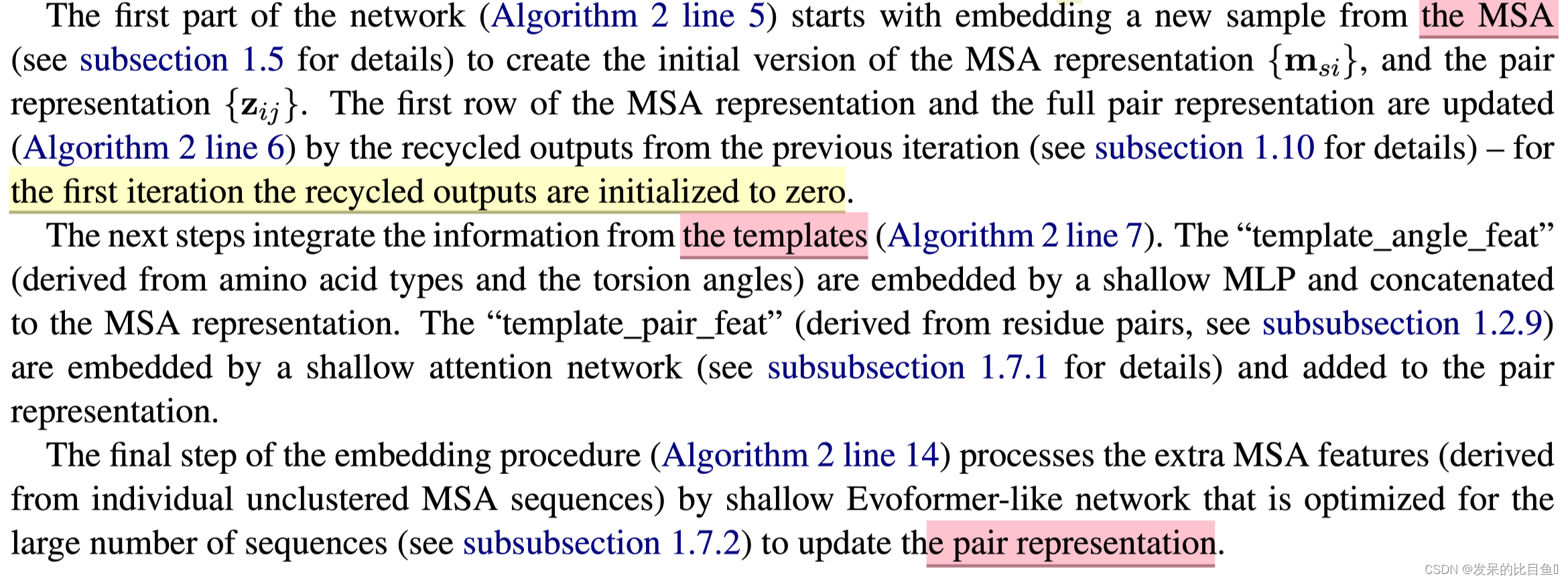
网络的第一部分首先从嵌入一个来自MSA的新示例开始,以创建MSA_{m_si}表示和pair_{z_ij}表示的初始版本。MSA表示的第一行和完整的对表示由来自前一个迭代的回收输出更新,对于第一个迭代,回收输出初始化为零。
Template Embedding 和 Pair Embedding:
接下来的步骤将集成来自模板的信息。template_angle_feat通过浅层MLP嵌入并连接到MSA表示。template_pair_feat由一个浅注意网络嵌入,并添加到pair表示中。
嵌入过程的最后一步通过浅Evoformer-like网络处理额外的MSA特征,该网络针对大量序列进行了优化,以更新pair表示。
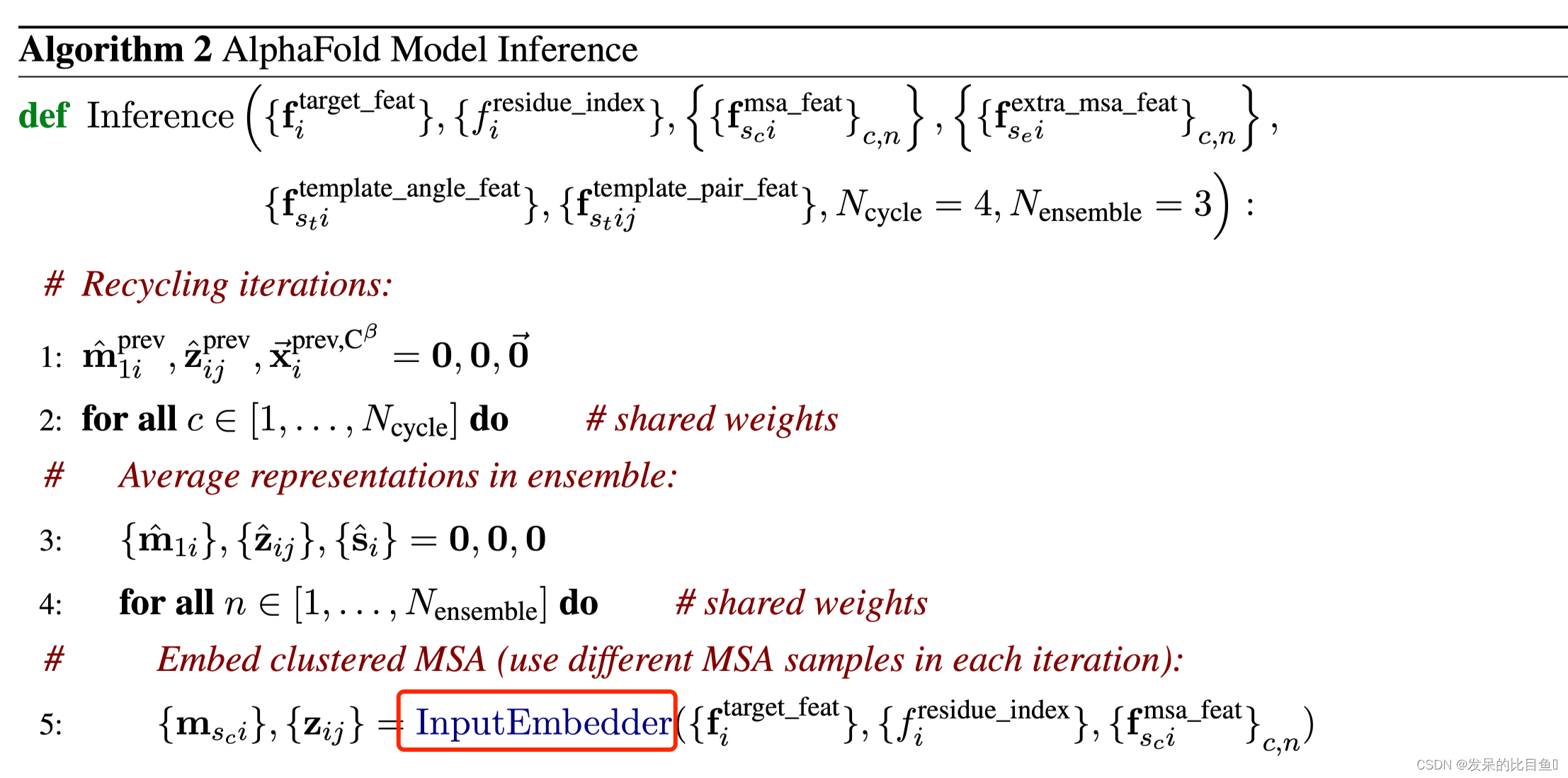
代码细节
MSA Representation
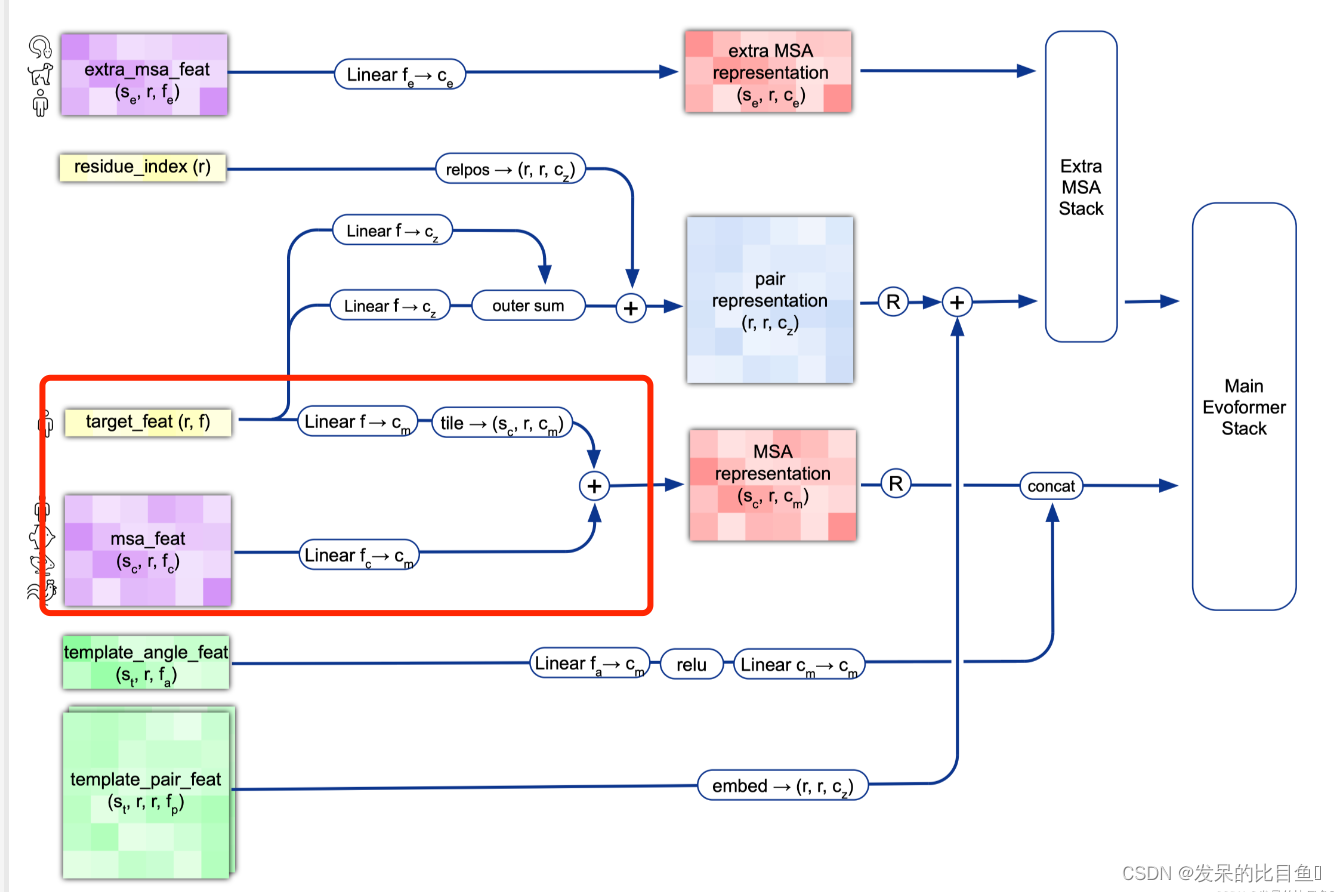
preprocess_1d = common_modules.Linear( # 初始化线性层c.msa_channel, name='preprocess_1d')( # c.msa_channel 256batch['target_feat']) #(84, 22) --> (84, 256)preprocess_msa = common_modules.Linear(c.msa_channel, name='preprocess_msa')(batch['msa_feat']) # (508, 84, 49) --> (508, 84, 256)msa_activations = jnp.expand_dims(preprocess_1d, axis=0) + preprocess_msa
Pair Representation
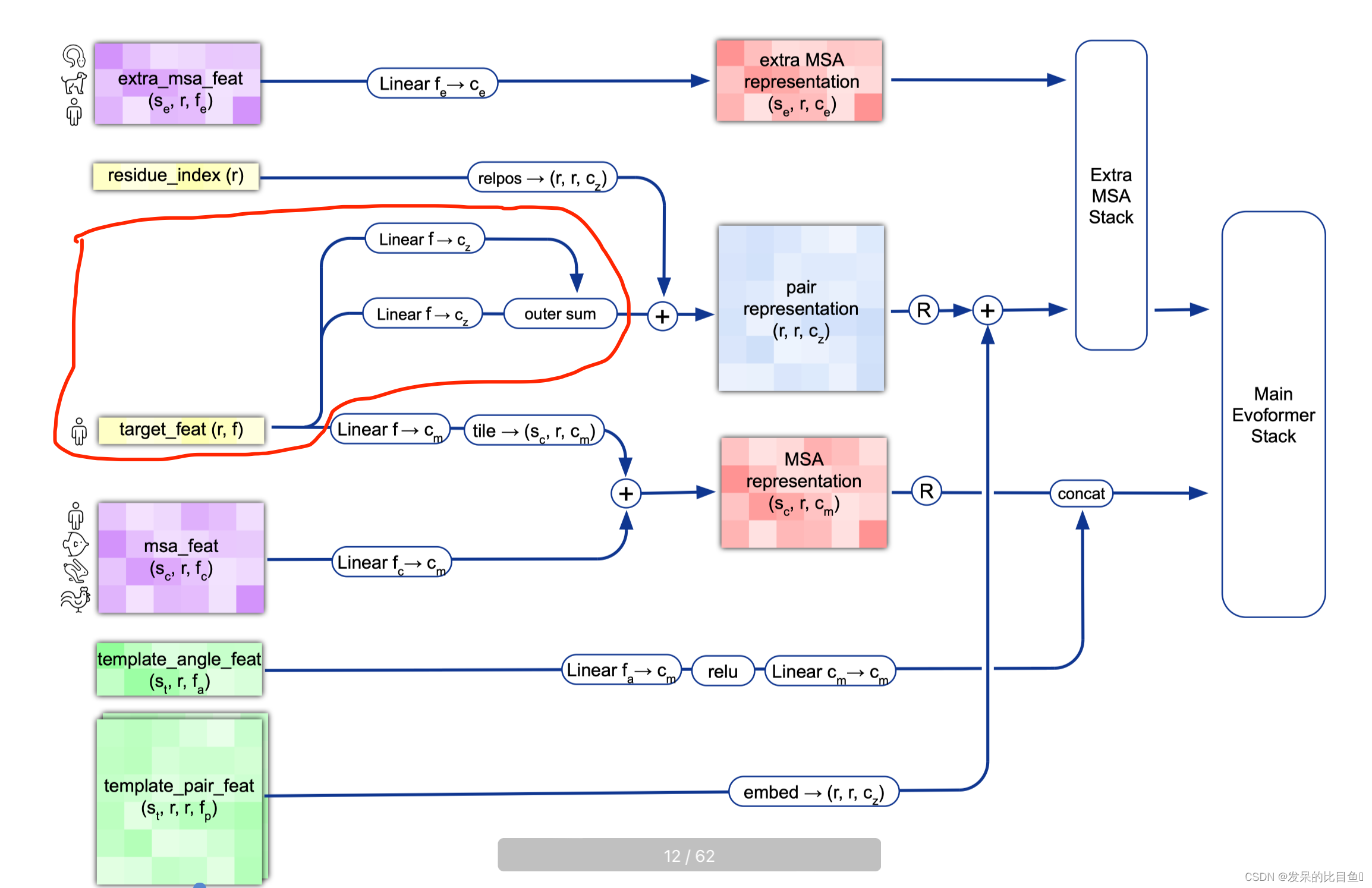
注入以前的输出进行回收
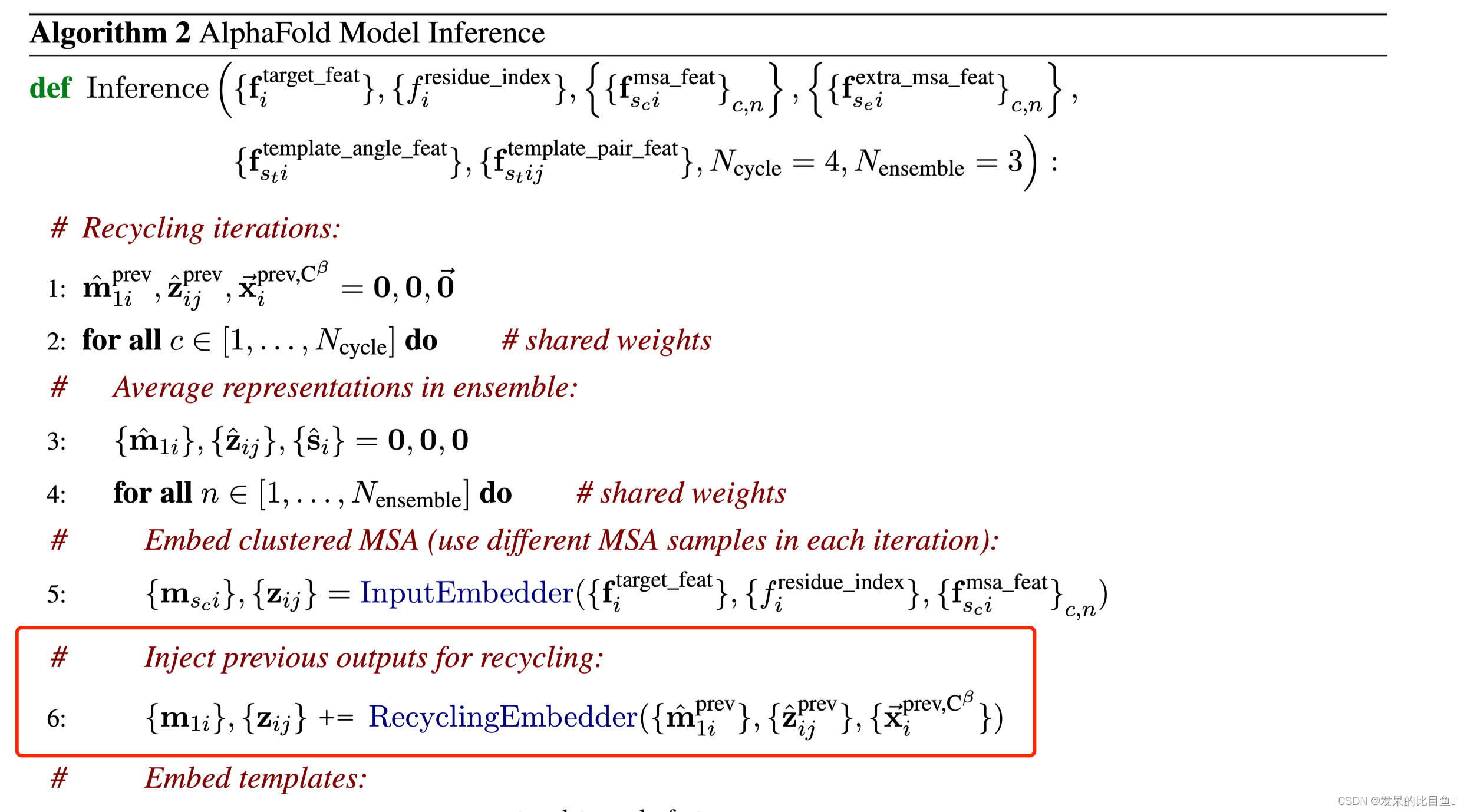
## 位置信息if c.recycle_pos: prev_pseudo_beta = pseudo_beta_fn(batch['aatype'], batch['prev_pos'], None) # (84, 3)dgram = dgram_from_positions(prev_pseudo_beta, **self.config.prev_pos)pair_activations += common_modules.Linear(c.pair_channel, name='prev_pos_linear')(dgram)# 特征信息if c.recycle_features: prev_msa_first_row = hk.LayerNorm(axis=[-1], create_scale=True, create_offset=True, name='prev_msa_first_row_norm')(batch['prev_msa_first_row']) # (84, 256) --> (84, 256) 取第一行MSAmsa_activations = msa_activations.at[0].add(prev_msa_first_row) # 第一行的加到msa_activations第一行## Pair 信息pair_activations += hk.LayerNorm( axis=[-1], create_scale=True, create_offset=True, name='prev_pair_norm')(batch['prev_pair']) ## (84, 84, 128) --> (84, 84, 128).......
def pseudo_beta_fn(aatype, all_atom_positions, all_atom_masks):"""Create pseudo beta features. 创建伪测试功能"""# (84, 37, 3) --> atom 空间位置信息is_gly = jnp.equal(aatype, residue_constants.restype_order['G']) # 是否是gly氨基酸ca_idx = residue_constants.atom_order['CA'] # C_α 索引 1cb_idx = residue_constants.atom_order['CB'] # C_β 索引 3pseudo_beta = jnp.where( # is_gly 1 选择 ca_idx 否则 选择 cb_idx --> (84, 3)jnp.tile(is_gly[..., None], [1] * len(is_gly.shape) + [3]), # 将函数沿着X或者Y轴扩大n倍,jnp.tile((84,1), [1,3]) -> (84, 3)all_atom_positions[..., ca_idx, :],all_atom_positions[..., cb_idx, :]) # all_atom_positions[..., cb_idx, :]--> (3,)if all_atom_masks is not None:pseudo_beta_mask = jnp.where(is_gly, all_atom_masks[..., ca_idx], all_atom_masks[..., cb_idx])pseudo_beta_mask = pseudo_beta_mask.astype(jnp.float32)return pseudo_beta, pseudo_beta_maskelse:return pseudo_betadef dgram_from_positions(positions, num_bins, min_bin, max_bin):"""Compute distogram from amino acid positions. 根据氨基酸位置计算距离图positions: [N_res, 3] Position coordinates. 位置:[N_res,3]位置坐标。num_bins: The number of bins in the distogram. num_bins:分布图中的箱数。min_bin: The left edge of the first bin. min_bin:第一个bin的左边缘。max_bin: The left edge of the final bin. The final bin catches max_bin:最终bin的左边缘。最后一个bin将捕获大于“max_bin”的"""def squared_difference(x, y):return jnp.square(x - y)lower_breaks = jnp.linspace(min_bin, max_bin, num_bins)lower_breaks = jnp.square(lower_breaks) # 下限(15)upper_breaks = jnp.concatenate([lower_breaks[1:],jnp.array([1e8], dtype=jnp.float32)], axis=-1) # 上限dist2 = jnp.sum(squared_difference(jnp.expand_dims(positions, axis=-2), # (84, 1, 3)jnp.expand_dims(positions, axis=-3)), # (1, 84, 3) ## 上下两部分正好是残基对相互匹配求差,axis=-1, keepdims=True) ##[84, 84, 1]dgram = ((dist2 > lower_breaks).astype(jnp.float32) *(dist2 < upper_breaks).astype(jnp.float32))return dgram ## 保留残基之间距离为〉lower_breaks 〈 upper_breaks , 这个是mask (84, 84, bin)-> 最后一维是不同桶的分布。。。。。。。
if c.recycle_features: # 特征信息prev_msa_first_row = hk.LayerNorm(axis=[-1],create_scale=True,create_offset=True,name='prev_msa_first_row_norm')(batch['prev_msa_first_row']) # (84, 256) --> (84, 256) 取第一行MSA
关联最大距离特征
if c.max_relative_feature: # 相互关联的最大距离# Add one-hot-encoded clipped residue distances to the pair activations.pos = batch['residue_index']offset = pos[:, None] - pos[None, :] # (84, 84) pair相对位置相减rel_pos = jax.nn.one_hot(jnp.clip(offset + c.max_relative_feature,a_min=0,a_max=2 * c.max_relative_feature),2 * c.max_relative_feature + 1) ## 位置差信息pair_activations += common_modules.Linear(c.pair_channel, name='pair_activiations')(rel_pos) # (84, 84, 65) -> (84, 84, 128)
模版特征
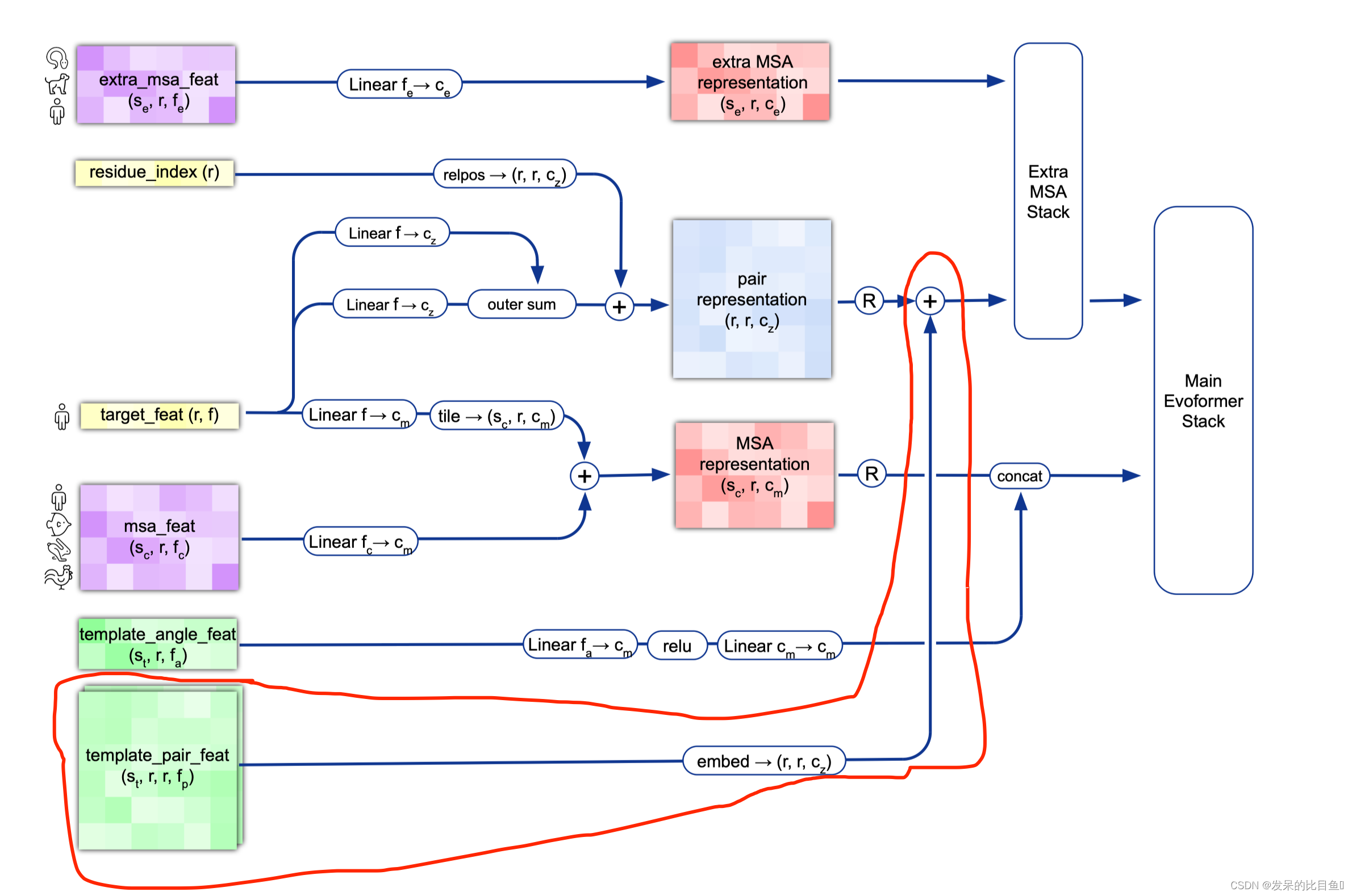
使用的预处理信息
[k for k in batch.keys() if k.startswith('template_')]
['template_aatype', # 氨基酸序列的one-hot表示 [N_temp, N_res, 22]
'template_all_atom_masks', # [N_temp, n_res, 37]
'template_all_atom_positions', #原子信息 [N_temp, n_res, 37, 3]
'template_mask', #[N-temp]
'template_pseudo_beta', #[N_temp, N_res, 3]
'template_pseudo_beta_mask', # [N_temp, N_res] 指示β-碳(甘氨酸的α-碳)原子是否具有该残基处模板的坐标的掩码
'template_sum_probs'] # [n_temp, 1]
if c.template.enabled: # 是否使用模版template_batch = {k: batch[k] for k in batch if k.startswith('template_')}template_pair_representation = TemplateEmbedding(c.template, gc)(pair_activations,template_batch,mask_2d,is_training=is_training)pair_activations += template_pair_representation........额外的MSA Representation
extra_msa_feat = create_extra_msa_feature(batch)
extra_msa_activations = common_modules.Linear(c.extra_msa_channel, name='extra_msa_activations')(extra_msa_feat)def create_extra_msa_feature(batch):"""将extra_msa扩展为1hot,并与其他额外的msa功能合并。
我们尽可能晚做这件事,因为一个小时的额外msa可能非常大。"""# 23 = 20 amino acids + 'X' for unknown + gap + bert maskmsa_1hot = jax.nn.one_hot(batch['extra_msa'], 23)msa_feat = [msa_1hot,jnp.expand_dims(batch['extra_has_deletion'], axis=-1),jnp.expand_dims(batch['extra_deletion_value'], axis=-1)]return jnp.concatenate(msa_feat, axis=-1)
Extra MSA Stack
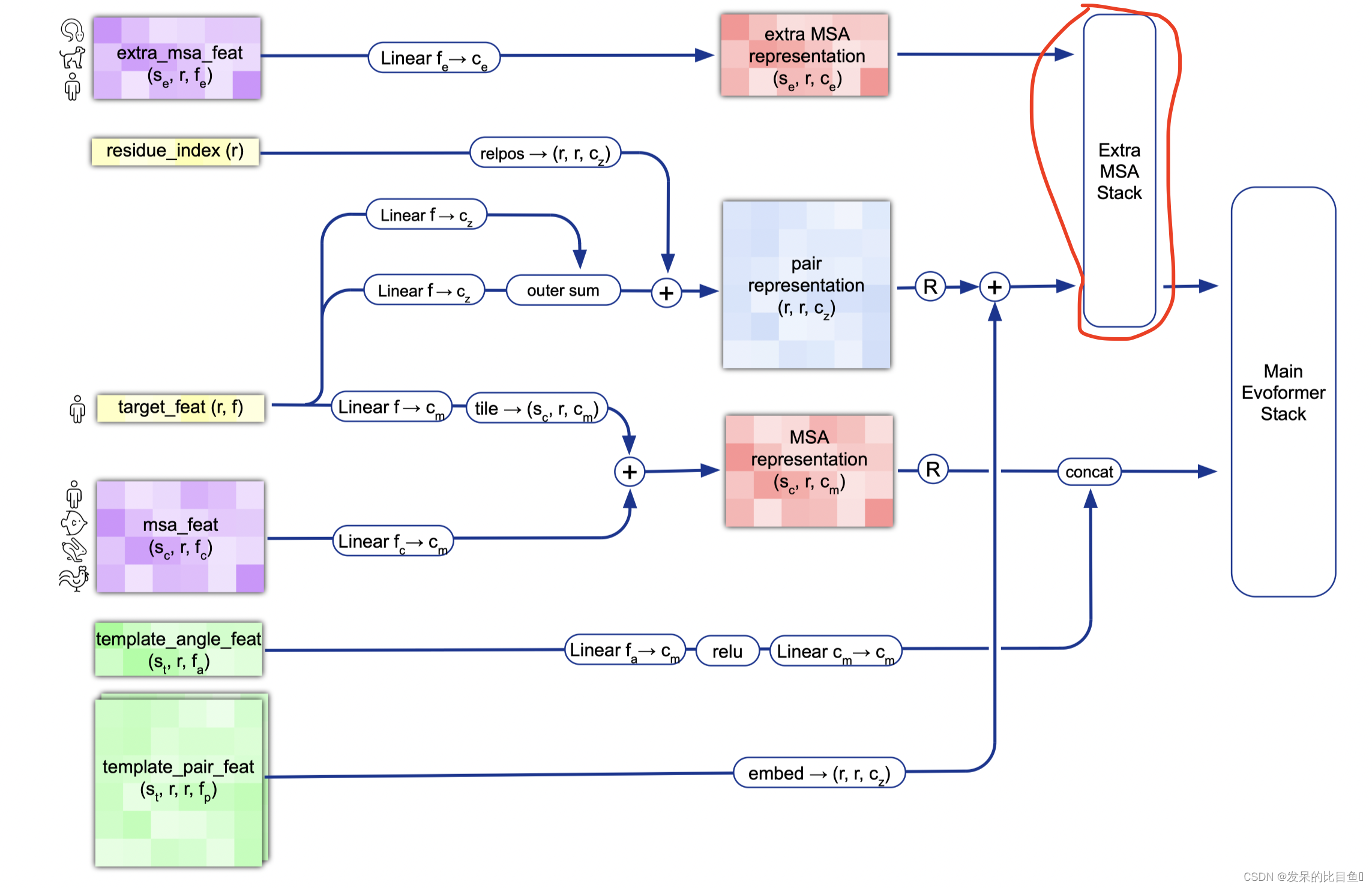
extra_msa_stack_input = {'msa': extra_msa_activations,'pair': pair_activations,}extra_msa_stack_iteration = EvoformerIteration(c.evoformer, gc, is_extra_msa=True, name='extra_msa_stack')def extra_msa_stack_fn(x):act, safe_key = xsafe_key, safe_subkey = safe_key.split()extra_evoformer_output = extra_msa_stack_iteration(activations=act,masks={'msa': batch['extra_msa_mask'],'pair': mask_2d},is_training=is_training,safe_key=safe_subkey)return (extra_evoformer_output, safe_key)if gc.use_remat:extra_msa_stack_fn = hk.remat(extra_msa_stack_fn)extra_msa_stack = layer_stack.layer_stack(c.extra_msa_stack_num_block)(extra_msa_stack_fn)extra_msa_output, safe_key = extra_msa_stack((extra_msa_stack_input, safe_key))pair_activations = extra_msa_output['pair']evoformer_input = {'msa': msa_activations,'pair': pair_activations,}
MSA 与模版角度特征concat
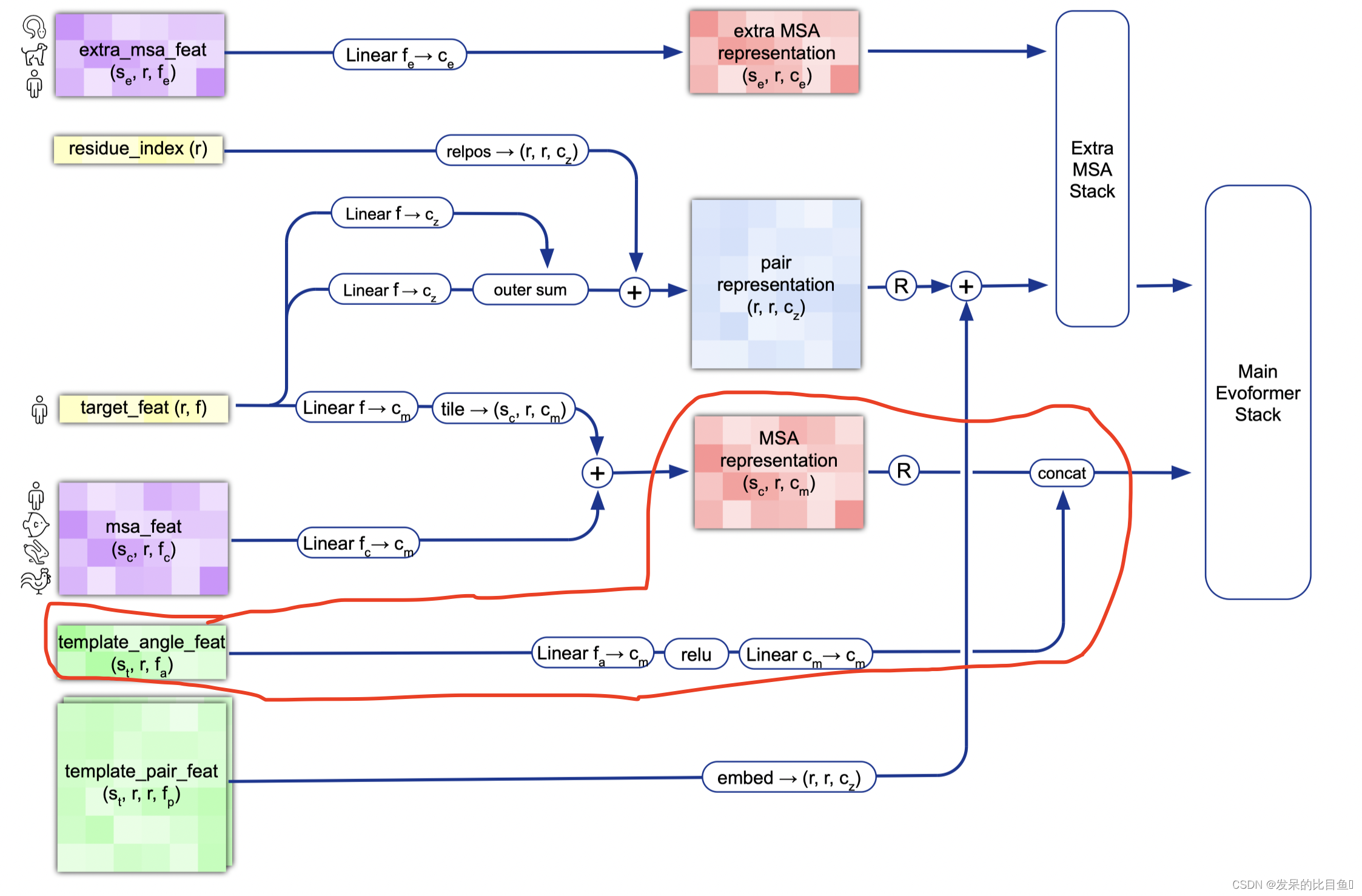
evoformer_masks = {'msa': batch['msa_mask'], 'pair': mask_2d}# Append num_templ rows to msa_activations with template embeddings.
# Jumper et al. (2021) Suppl. Alg. 2 "Inference" lines 7-8
if c.template.enabled and c.template.embed_torsion_angles:num_templ, num_res = batch['template_aatype'].shape# Embed the templates aatypes.aatype_one_hot = jax.nn.one_hot(batch['template_aatype'], 22, axis=-1)# Embed the templates aatype, torsion angles and masks.# Shape (templates, residues, msa_channels)ret = all_atom.atom37_to_torsion_angles(aatype=batch['template_aatype'],all_atom_pos=batch['template_all_atom_positions'],all_atom_mask=batch['template_all_atom_masks'],# Ensure consistent behaviour during testing:placeholder_for_undefined=not gc.zero_init)template_features = jnp.concatenate([aatype_one_hot,jnp.reshape(ret['torsion_angles_sin_cos'], [num_templ, num_res, 14]),jnp.reshape(ret['alt_torsion_angles_sin_cos'], [num_templ, num_res, 14]),ret['torsion_angles_mask']], axis=-1)template_activations = common_modules.Linear(c.msa_channel,initializer='relu',name='template_single_embedding')(template_features)template_activations = jax.nn.relu(template_activations)template_activations = common_modules.Linear(c.msa_channel,initializer='relu',name='template_projection')(template_activations)# Concatenate the templates to the msa.evoformer_input['msa'] = jnp.concatenate([evoformer_input['msa'], template_activations], axis=0)# Concatenate templates masks to the msa masks.# Use mask from the psi angle, as it only depends on the backbone atoms# from a single residue.torsion_angle_mask = ret['torsion_angles_mask'][:, :, 2]torsion_angle_mask = torsion_angle_mask.astype(evoformer_masks['msa'].dtype)evoformer_masks['msa'] = jnp.concatenate([evoformer_masks['msa'], torsion_angle_mask], axis=0)
网络主干线evoformer
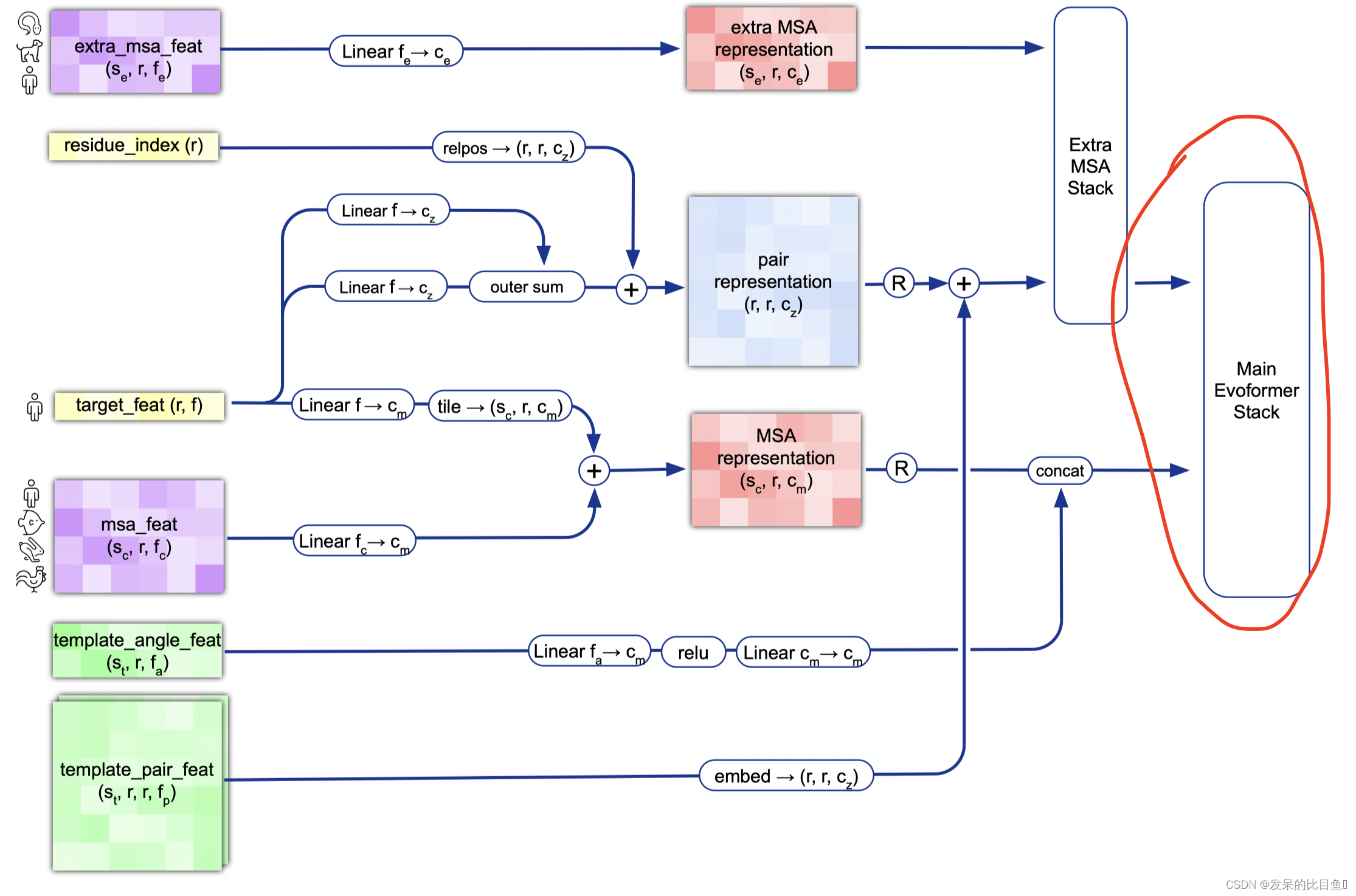
evoformer_iteration = EvoformerIteration(c.evoformer, gc, is_extra_msa=False, name='evoformer_iteration')
EvoformerIteration网络的处理过程比较复杂, 我们这里省略,这里只讲解该网络的输入和输出。
相关内容
热门资讯
汽车油箱结构是什么(汽车油箱结...
本篇文章极速百科给大家谈谈汽车油箱结构是什么,以及汽车油箱结构原理图解对应的知识点,希望对各位有所帮...
美国2年期国债收益率上涨15个...
原标题:美国2年期国债收益率上涨15个基点 美国2年期国债收益率上涨15个基...
嵌入式 ADC使用手册完整版 ...
嵌入式 ADC使用手册完整版 (188977万字)💜&#...
重大消息战皇大厅开挂是真的吗...
您好:战皇大厅这款游戏可以开挂,确实是有挂的,需要了解加客服微信【8435338】很多玩家在这款游戏...
盘点十款牵手跑胡子为什么一直...
您好:牵手跑胡子这款游戏可以开挂,确实是有挂的,需要了解加客服微信【8435338】很多玩家在这款游...
senator香烟多少一盒(s...
今天给各位分享senator香烟多少一盒的知识,其中也会对sevebstars香烟进行解释,如果能碰...
终于懂了新荣耀斗牛真的有挂吗...
您好:新荣耀斗牛这款游戏可以开挂,确实是有挂的,需要了解加客服微信8435338】很多玩家在这款游戏...
盘点十款明星麻将到底有没有挂...
您好:明星麻将这款游戏可以开挂,确实是有挂的,需要了解加客服微信【5848499】很多玩家在这款游戏...
总结文章“新道游棋牌有透视挂吗...
您好:新道游棋牌这款游戏可以开挂,确实是有挂的,需要了解加客服微信【7682267】很多玩家在这款游...
终于懂了手机麻将到底有没有挂...
您好:手机麻将这款游戏可以开挂,确实是有挂的,需要了解加客服微信【8435338】很多玩家在这款游戏...